Research
Lighting Up the Central Dogma in Embryonic Development
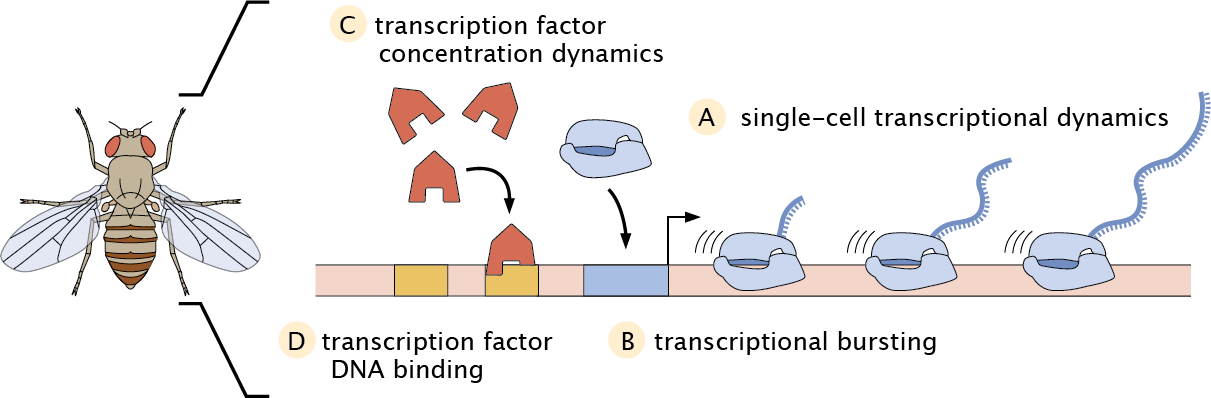
In our lab, we develop new experimental and computational tools to measure and manipulate the flow of information along the central dogma in single cells of living, developing embryos using the fruit fly Drosophila melanogaster as a case study.
ALighting up transcriptional dynamics using MS2
We have pioneered the MS2/MCP technology in developing fruit fly embryos, which allows for live-cell imaging of nascent RNA transcription with single-cell resolution.
Video: A developing Drosophila embryo with labeled histones marking the nuclei (red) and MS2-labelled nascent transcripts of the even-skipped gene (green),
which forms seven stripes by the end of nuclear cycle 14 (nc14).
Relevant Publications: Garcia et al. (2013). Bothma et al. (2014).
BLighting up the transcription cycle using computational methods
We employ powerful computational inference methods to extract information about the transcription cycle from our experimental MS2 data such as bursting kinetics and RNAP elongation rates.
Video: Active (green) and inactive (red) nuclei transcribing even-skipped (eve) stripe 2 computationally identified using a Hidden Markov Model.
Relevant Publications: Lammers et. al. (2020). Berrocal et. al. (2020). Liu et. al. (2020).
CLighting up transcription factor dynamics using LlamaTags
Slowly maturing fluorescent fusions are insufficient to reveal transcription factor concentration dynamics in development. To circumvent this challenge, we have developed the LlamaTag technology, a technique for rapid fluorescent labeling of newly expressed proteins that allows for single-cell, live imaging of protein expression at high spatiotemporal resolution.
Video: A developing Drosophila embryo with MS2-labelled nascent mRNA transcripts (red) and LlamaTag-labeled proteins (green)
of the transcription factor fushi tarazu (ftz).
Relevant Publications: Bothma et. al. (2018).
DLighting up transcription factor binding dynamics
Using cutting-edge lattice light sheet microscopy (with Xavier Darzacq, UC Berkeley), we have demonstrated the detection of the binding of individual activator molecules to the DNA in living embryos, and have discovered localized “hubs” of enriched binding throughout the nucleus.
Video: Individual molecules of the maternal activator Bicoid bind in the nuclei of a developing Drosophila embryo.
Relevant Publications: Mir et. al. (2017).
Lighting Up The Central Dogma Beyond Flies
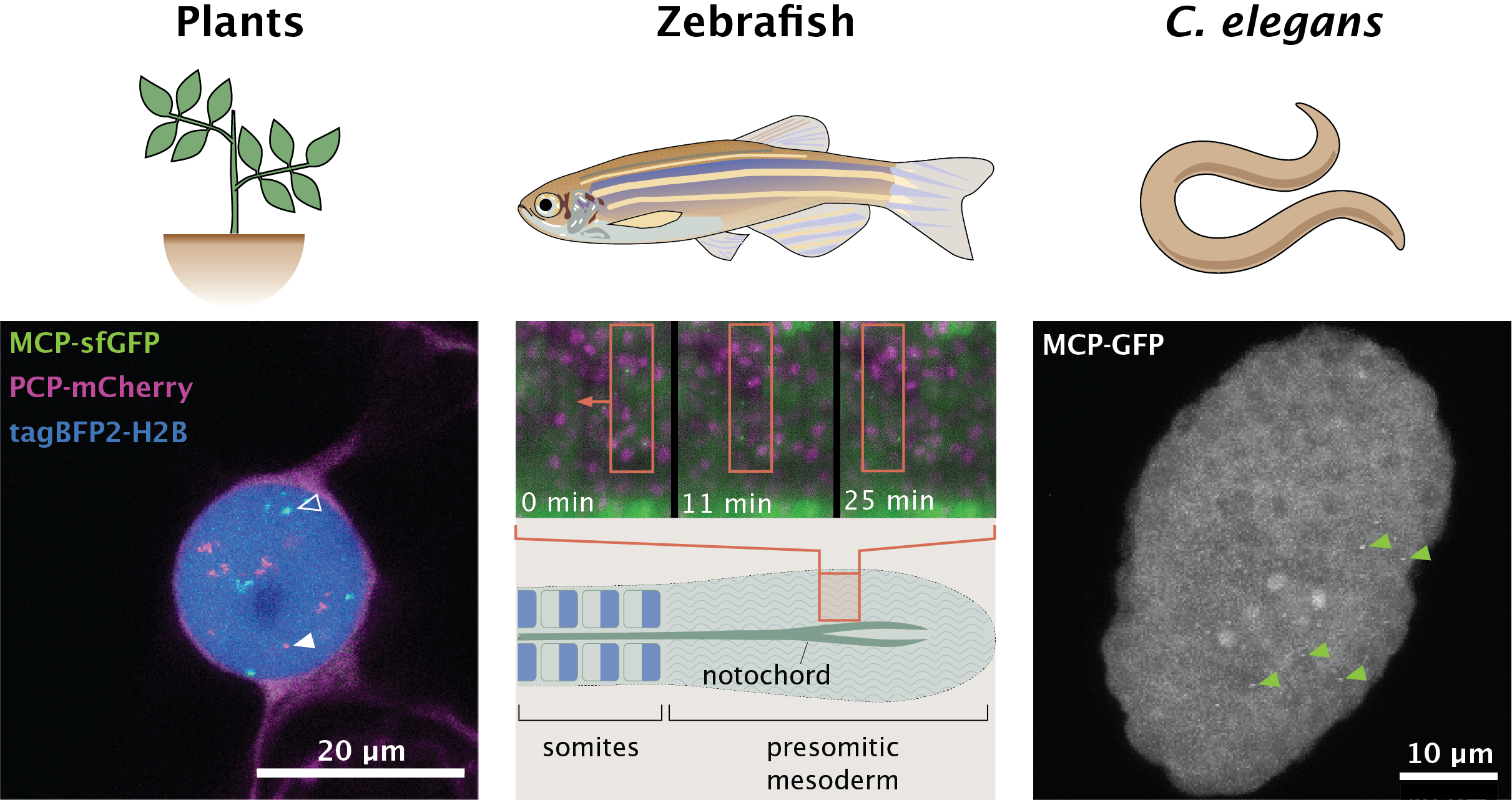
Image: (left) A tobacco leaf epidermal cell expressing BFP-tagged histones and nascent mRNA transcripts from two reporter constructs tagged with PP7 (magenta, closed arrowhead) or MS2 (green, open arrowhead).
(center) A zebrafish embryo expressing mKate-tagged histones (magenta) and MS2-tagged her1 transcripts (green). Note the movement of her1-expressing cells within the presomitic mesoderm as embryogenesis progresses.
(right) A C. elegans embryo expressing nascent mRNA transcripts labelled with MS2 stem loops (green arrows point to a subset of MS2 puncta).
Relevant Publications: Alamos et. al. (2021).
Physical Biology of Living Embryos
Using our technologies to quantify the processes of the central dogma, we are engaged in a dialogue between theory and experiment aimed at reaching a predictive understanding of developmental decision-making.Bending nature to understand it: Using synthetic biology to derive governing equations of transcriptional control in development
Writing theoretical models of endogenous regulatory regions in development calls for governing equations with too many unknown parameters. As an alternative approach to modeling endogenous complexity, we have developed several synthetic enhancer systems that attempt to build eukaryotic transcription from the ground up. The ensuing models are much more tractable, making it possible to engage in a dialogue between theory and experiment to test our theoretical predictions.
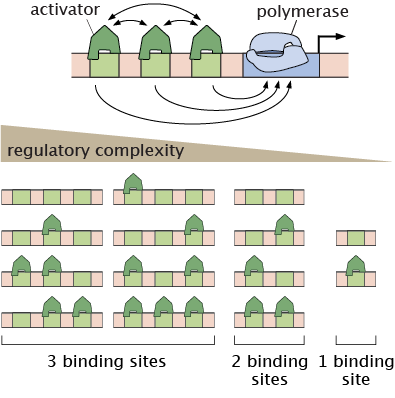
Image: Multiplicity of promoter occupancy states for one, two, and three TF binding site architectures.
Relevant Publications: Garcia et. al. (2016). Reimer et. al. (2021). Kim et. al. (2021).
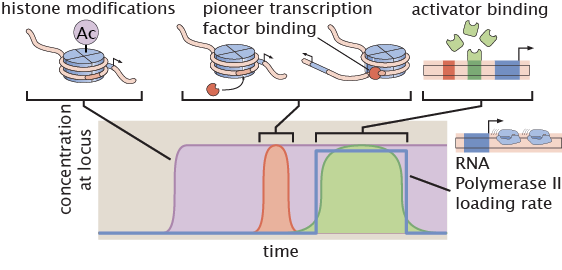
Regulation of DNA accessibility
Access to DNA constitutes an important regulatory layer in eukaryotic gene regulation. With our ability to resolve transcription in time and space, we are probing the predictive power of models of DNA accessibility regulation both in and out of thermodynamic equilibrium.
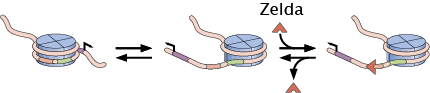
Relevant Publications: Eck et. al. (2020).
Anatomy of a transcriptional burst
Transcriptional bursting seems to be more the rule than the exception throughout biology. Yet, while we have uncovered which bursting parameters (frequency, duration, and amplitude) are controlled by transcription factors, we remain ignorant about how this control is implemented at the molecular level. With our ability to characterize both transcription factor input and transcriptional output dynamics at the single cell level, we are developing new analysis methods to reveal the Anatomy of a Burst: the precise ordering of biochemical events that occur prior, during, and after a burst, at individual promoters in living embryos with the time resolution of a few seconds.
Relevant Publications: Lammers et. al. (2020).